Archaeologists Discover Oldest Direct Evidence for Honey Collecting in Africa in Ancient Clay Pots
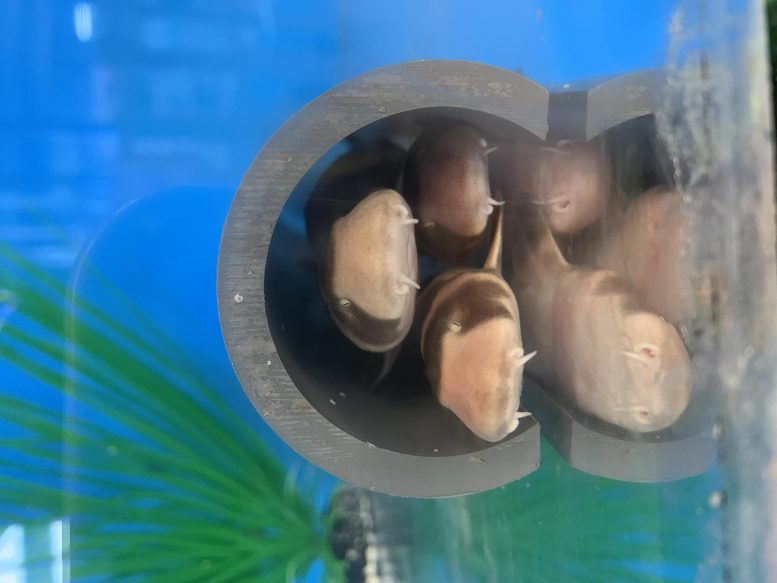
Traces of beeswax were detected in 3500 year-old clay pots like this. Credit: Peter Breunig, Goethe University Frankfurt
Scientists at Goethe University and University of Bristol (UK) find traces of beeswax in prehistoric pottery of the West African Nok culture.
Before sugar cane and sugar beets conquered the world, honey was the worldwide most important natural product for sweetening. Archaeologists at Goethe University in cooperation with chemists at the University of Bristol have now produced the oldest direct evidence of honey collecting of in Africa. They used chemical food residues in potsherds found in Nigeria.
Honey is humankind’s oldest sweetener – and for thousands of years it was also the only one. Indirect clues about the significance of bees and bee products are provided by prehistoric petroglyphs on various continents, created between 8,000 and 40,000 years ago. Ancient Egyptian reliefs indicate the practice of beekeeping as early as 2600 year BCE. But for sub-Saharan Africa, direct archaeological evidence has been lacking until now. The analysis of the chemical residues of food in potsherds has fundamentally altered the picture. Archaeologists at Goethe University in cooperation with chemists at the University of Bristol were able to identify beeswax residues in 3500 year-old potsherds of the Nok culture.
The Nok culture in central Nigeria dates between 1500 BCE and the beginning of the Common Era and is known particularly for its elaborate terracotta sculptures. These sculptures represent the oldest figurative art in Africa. Until a few years ago, the social context in which these sculptures had been created was completely unknown. In a project funded by the German Research Foundation, Goethe University scientists have been studying the Nok culture in all its archaeological facets for over twelve years. In addition to settlement pattern, chronology, and meaning of the terracotta sculptures, the research also focussed on environment, subsistence and diet.
Did the people of the Nok Culture have domesticated animals or were they hunters? Archaeologists typically use animal bones from excavations to answer these questions. But what to do if the soil is so acidic that bones are not preserved, as is the case in the Nok region?
The analysis of molecular food residues in pottery opens up new possibilities. This is because the processing of plant and animal products in clay pots releases stable chemical compounds, especially fatty acids (lipids). These can be preserved in the pores of the vessel walls for thousands of years, and can be detected with the assistance of gas chromatography.
To the researchers’ great surprise, they found numerous other components besides the remains of wild animals, significantly expanding the previously known spectrum of animals and plants used. There is one creature in particular that they had not expected: the honeybee. A third of the examined shards contained high-molecular lipids, typical for beeswax.
It is not possible to reconstruct from the lipids which bee products were used by the people of the Nok culture. Most probably they separated the honey from the waxy combs by heating them in the pots. But it is also conceivable that honey was processed together with other raw materials from animals or plants, or that they made mead. The wax itself could have served technical or medical purposes. Another possibility is the use of clay pots as beehives, as is practiced to this day in traditional African societies.
“We began this study with our colleagues in Bristol because we wanted to know if the Nok people had domesticated animals,” explains Professor Peter Breunig from Goethe University, who is the director of the archaeological Nok project. “That honey was part of their daily menu was completely unexpected, and unique in the early history of Africa until now.”
Dr. Julie Dunne from the University of Bristol, first author of the study says: “This is a remarkable example of how biomolecular information from prehistoric pottery in combination with ethnographic data provides insight into the use of honey 3500 years ago.”
Professor Richard Evershed, Head of the Institute for Organic Chemistry at the University of Bristol and co-author of the study points out that the special relationship between humans and honeybees was already known in antiquity. “But the discovery of beeswax residues in Nok pottery allows a very unique insight into this relationship, when all other sources of evidence are lacking.”
Professor Katharina Neumann, who is in charge of archaeobotany in the Nok project at Goethe University says: “Plant and animal residues from archaeological excavations reflect only a small section of what prehistoric people ate. The chemical residues make previously invisible components of the prehistoric diet visible.” The first direct evidence of beeswax opens up fascinating perspectives for the archaeology of Africa. Neumann: “We assume that the use of honey in Africa has a very long tradition. The oldest pottery on the continent is about 11,000 years old. Does it perhaps also contain beeswax residues? Archives around the world store thousands of ceramic shards from archaeological excavations that are just waiting to reveal their secrets through gas chromatography and paint a picture of the daily life and diet of prehistoric people.”
For more on this research, read Ancient Pottery Reveals First Evidence of Prehistoric Honey Hunting in West Africa 3,500 Years Ago.
Reference: “Honey-collecting in prehistoric West Africa from 3,500 years ago” by Julie Dunne, Alexa Höhn, Gabriele Franke, Katharina Neumann, Peter Breunig, Toby Gillard, Caitlin Walton-Doyle and Richard P. Evershed, 14 April 2021, Nature Communications.
DOI: 10.1038/s41467-021-22425-4